Agilent Microarray File Format

Jump to:, For two-channel analysis, refer to. Note/Disclaimer: The instructions on this page are provided 'as-is'. They worked for my application and have helped many others on their way. Launch R and load in the LIMMA library. The LIMMA user's guide, citation information, downloads and installation information is available from the.
Library(limma) 2. Read in your tab-delimited targets file, which contains both the names of your Agilent Feature Extraction raw data text files and the corresponding sample information. To describe 6 arrays that have been hybridised with two biological replicates of three different tissues, the format of the file should look something like this: SampleNumberFileNameCondition 1US83444010018S01GE1-v510Apr0812.txtBrain 2US83444010020S01GE1-v510Apr0811.txtBrain 3US83444010021S01GE1-v510Apr0811.txtLung 4US83444010021S01GE1-v510Apr0812.txtLung 5US83444010023S01GE1-v510Apr0811.txtLiver 6US83444010023S01GE1-v510Apr0812.txtLiver targets.
- Importing Data from Agilent’s Feature Extraction. Intensity information from the Agilent microarray. Selected for import will be moved to the Agilent Files.
- Mutually add a tab in the beggining of the header to make it a valid RMA format., I have started agilent microarray data. I can read soft files in R using.
File Format Converter
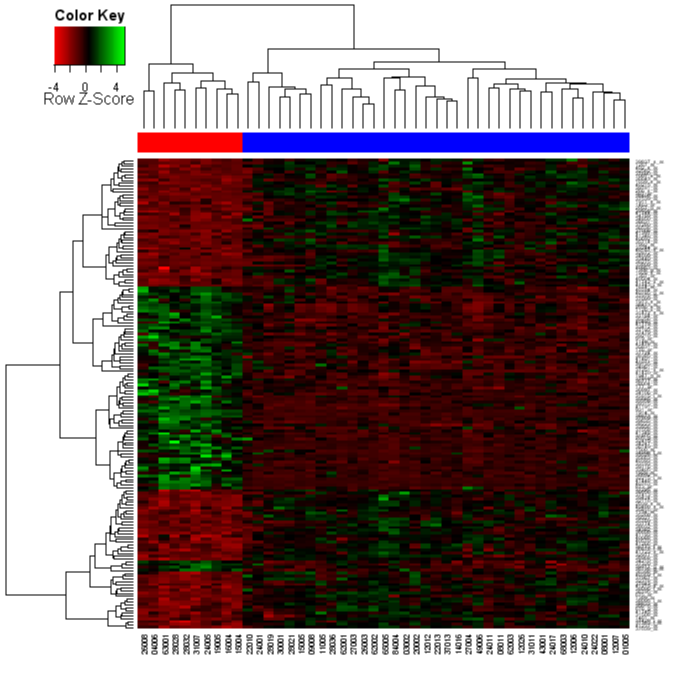
Analyzing Agilent microarray with MeV. I’m analyzing one-color Agilent chips data (human 44k). (Select filter loader/Other format files/Agilent files).